|
Question 1: For measuring the molecular mass of large organic molecules (eg. 40,000 Daltons), what is the accuracy of current mass spectrometers?
(Click on the radio-button at the left of the correct answer below.)
|
|
|
|
Your Answer: YourAnswer Mark
Correct Answer: Within 0.01 % of the total molecular mass of the sample (ie. 4 Daltons for a 40,000 Dalton molecule)..
For large molecules, current mass spectrometers can measure masses to an accuracy of 0.01% of the total molecular mass of the sample ie. within a 4 Daltons (Da) for a 40,000 Dalton molecule. This is sufficient to allow minor mass changes to be detected, eg. the substitution of one amino acid for another, or a post-translational modification.
For small organic molecules the molecular mass can be measured to within an accuracy of 5 ppm (equivalent to 0.0005 %) or less, which is often sufficient to confirm the molecular formula of a compound, and is also a standard requirement for publication in a chemical journal.
Structural information can be generated using certain types of mass spectrometers, usually those with multiple analysers which are known as tandem mass spectrometers. This is achieved by fragmenting the sample inside the instrument and analysing the products generated. This procedure is useful for the structural elucidation of organic compounds and for peptide or oligonucleotide sequencing.
|
|
|
Question 2: Which of the following components would not be found in a typical mass spectrometer?
(Click on the radio-button at the left of the correct answer below.)
|
|
|
|
Your Answer: YourAnswer Mark
Correct Answer: A Super-conducting magnet..
A mass spectrometer requires an ionization source, which may be Matrix-Assisted Laser Desorption/Ionization (MALDI), a Mass analyser which may use a Quadruploe to filter the ions, and a Detector, such as a Photo-multiplier. A vacuum pump is required to maintain a low pressure throughout the mass spectrometer to permit easy flow of the ions.
A Super-conducting magnet would be part of a NMR (Nuclear magnetic resonance) machine, not a typical Mass spectrometer, (although new more expensive 'Fourier Transform Ion Cyclotron Resonance, FT-ICR' mass spectrometers do contain a super-conducting magnet in their cyclotron).
|
|
|
Question 3: Which of the following is actually measured by a mass spectrometer?
|
|
|
|
Your Answer: YourAnswer Mark
Correct Answer: Mass-to-charge ratio of the sample ions..
A mass spectrometer measures the mass-to-charge (m/z) ratio of the sample ions. From this the mass of the sample ions can be calculated.
It is an NMR (Nuclear magnetic resonance) machine that records variations nuclear spin of the sample ions.
|
|
|
Question 4: The m/z spectrum below is from a mass-spectrometer using position-ionization (ie. protonated molecular ions).
|
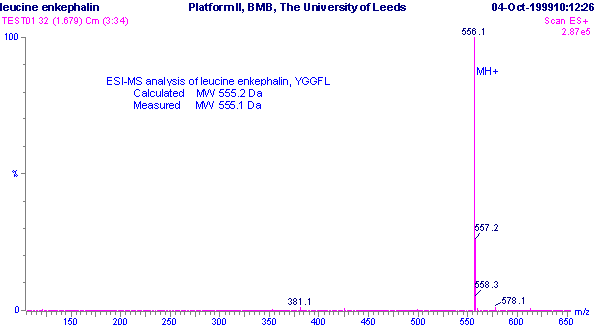
Positive ESI-MS m/z spectrum of leucine enkaphalin, YGGFL.
|
From this spectrum, what is the mass of the molecule being studied?
|
|
|
|
Your Answer: YourAnswer Mark
Correct Answer: 555.1 Daltons.
In Electro-spracy ionization, samples with molecular masses under 1200 Da tend to produce singly-charged ions, so in positive ionisation mode would have formula (M+H)+, where 'M' is the sample molecule and 'H' is a proton. So the mass of the molecule here will be position of the peak (556.1 Daltons) minus the mass of 1 proton (1 Dalton), giving 555.1 Daltons.
Protein masses are given in Daltons (also called the 'unified mass unit'), where one Dalton is approximately the mass of one hydrogen atom, or more precisely is one twelfth of an unbound carbon-12 atom.
|
|
|
Question 5: From the following molecular mass profile, what is the mass of the molecule being studied here?
|
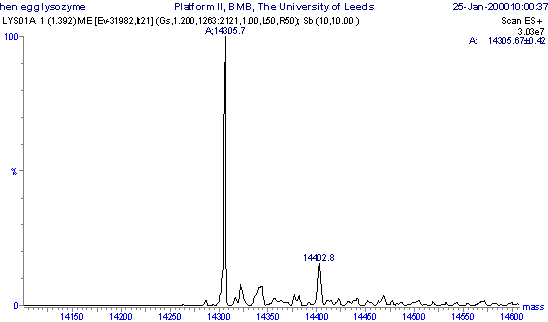
Molecular mass profile of lysozyme obtained by maximum entropy processing of the m/z spectrum.
|
|
|
|
|
Your Answer: YourAnswer Mark
Correct Answer: 14305.7 Daltons.
The molecular mass can be read directly from a Molecular mass profile. The tallest peak is at 14305.7 Daltons. The smaller peaks to the right are due to salt adducting, such as Na (M+23), K (M+39), H2SO4 or H3PO4 (M+98), where 'M' is the mass of the molecule.
This Molecular mass profile would have been obtained from the original m/z spectrum (shown below) by computer software using a method called 'maximum entropy processing'. Each of the peaks in the m/z spectrum below has a different number of protons (positive charges) attached to the sample molecule, so giving a different mass/charge (m/z) ratio. So: m/z = (MW + nH+)/n where: m/z = the mass-to-charge ratio; MW = the molecular mass of the sample; n = the integer number of charges on the ions; H = the mass of a proton = 1.008 Da. Using simulations equations between pairs of neighbouring peaks, the variable 'n' can be eliminated to calculate MW.
See 'section 5.3 Data processing' of An Introduction to Mass Spectrometry for more details.
|
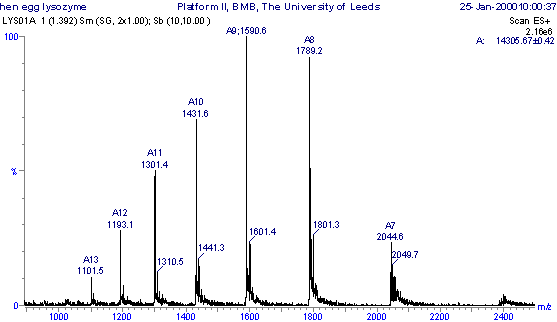
Positive ESI-MS m/z spectrum of the protien hen egg white lysozyme.
|
|